Antimicrobial resistance (AMR) is one of the most pressing global health challenges, driven by the rapid adaptation of bacterial pathogens to existing antibiotics and a stagnating discovery pipeline for new drugs. Our research addresses this gap by combining advanced bioinformatics, functional genomics, and physiologically relevant infection models to systematically identify novel, high-value drug targets and therapeutic strategies that are invisible to traditional discovery approaches.
Rather than relying on growth inhibition in artificial laboratory media, our work focuses on understanding how pathogens survive, adapt, and cause disease within the host environment. By interrogating bacterial genomes, transcriptomes, and fitness landscapes under infection-relevant conditions, we uncover conditionally essential genes and pathways that are critical for survival in vivo but dispensable under standard laboratory conditions. These pathways represent a largely untapped reservoir of innovative antimicrobial targets.
From big data to drug targets: Bioinformatics-driven discovery
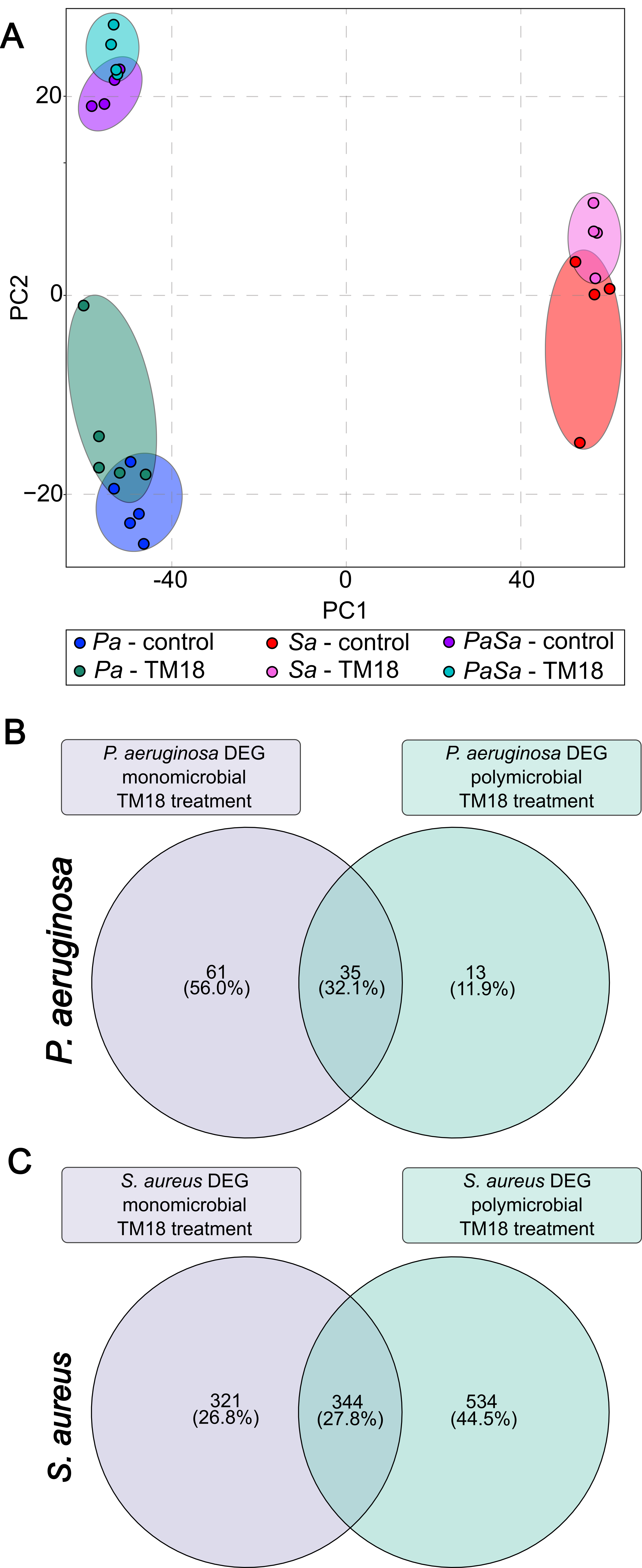
We leverage large-scale genomic and transcriptomic datasets, combined with state-of-the-art computational analysis, to map the genetic determinants of bacterial survival, virulence, and antibiotic resistance. Machine learning and systems-level approaches enable us to integrate diverse data types—including whole-genome sequencing, transposon sequencing (Tn-Seq), RNA-Seq, and phenotypic data—to identify previously unappreciated vulnerabilities in major pathogens such as Pseudomonas aeruginosa.
Functional genomics in host-relevant conditions
A key innovation of our programme is the application of functional genomics under physiologically relevant conditions, including host-mimicking media and murine infection models. Using Tn-Seq, we identify genes that are required specifically for survival in these environments, uncovering pathways involved in nutrient acquisition, stress adaptation, metabolic flexibility, and global regulation.
This strategy has revealed that many genes traditionally overlooked in antibiotic discovery—such as those involved in nucleotide metabolism, vitamin biosynthesis, and global stress responses—are essential for in vivo survival and virulence. Importantly, these pathways are often poorly targeted by existing antibiotics, making them attractive candidates for next-generation therapeutics.
Targeting global regulators and adaptive pathways
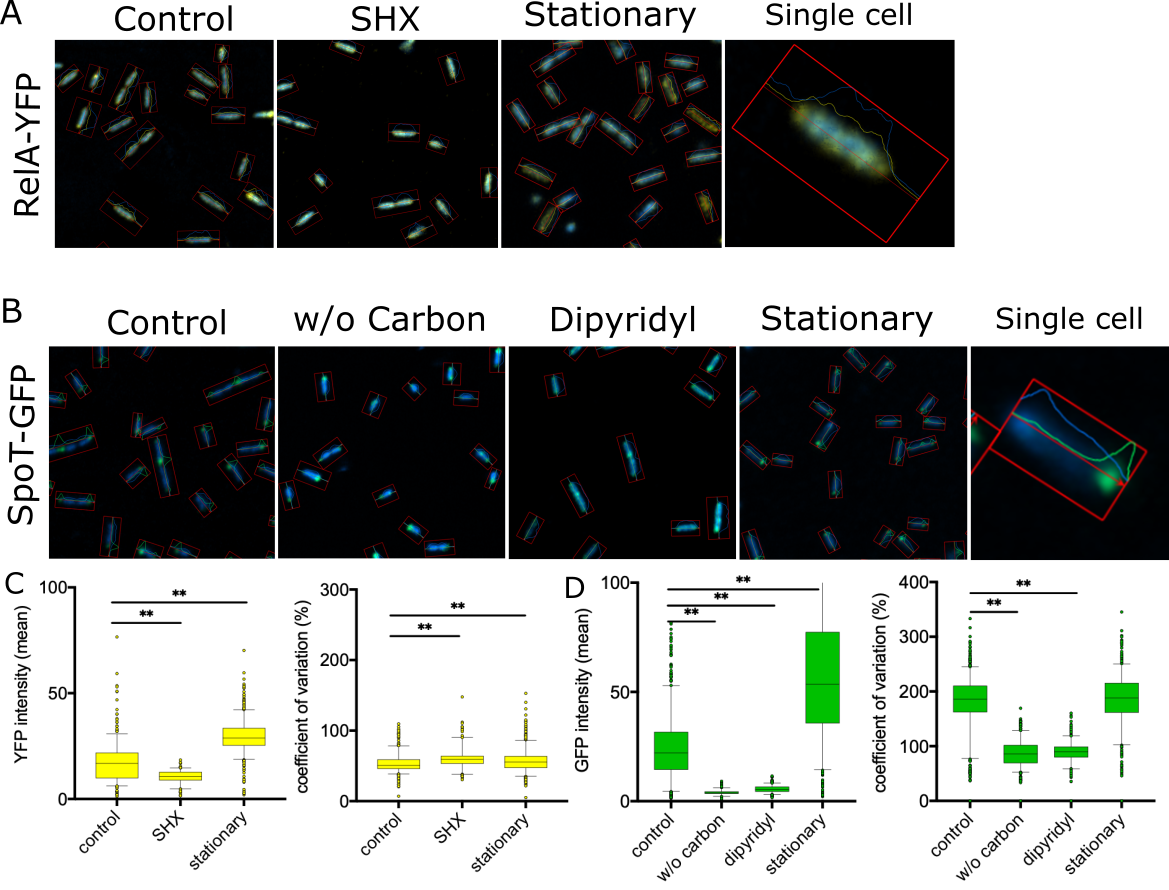
Our work also dissects global regulatory systems, such as the bacterial stringent stress response, that coordinate metabolism, virulence, and antibiotic tolerance. These systems act as central control hubs, allowing bacteria to adapt to hostile host environments and antimicrobial pressure rapidly. Disrupting such regulators can attenuate virulence, sensitise bacteria to existing drugs, and reduce the emergence of resistance, rather than simply killing bacteria outright.
These approaches enable us to:
- Predict novel resistance mechanisms and resistance-associated genes
- Identify metabolic and regulatory bottlenecks essential during infection
- Prioritise drug targets with reduced likelihood of resistance development
- Accelerate the transition from genomic data to experimentally validated candidates